Tutorial
Tutorial
Input Format
Input Format
| Prediction Engines | Data Columns | Extension/Input |
|---|---|---|
| Odorant Predictor | SMILES | *.csv |
| OR Finder | SMILES | *.csv |
| Odor Finder | Header of FASTA file, Receptor Sequence | *.csv |
| Odorant-OR Pair Analysis | SMILES, Header of FASTA file, Receptor Sequence | *.csv |
Output Format
Output Format
| Columns | Explanation |
|---|---|
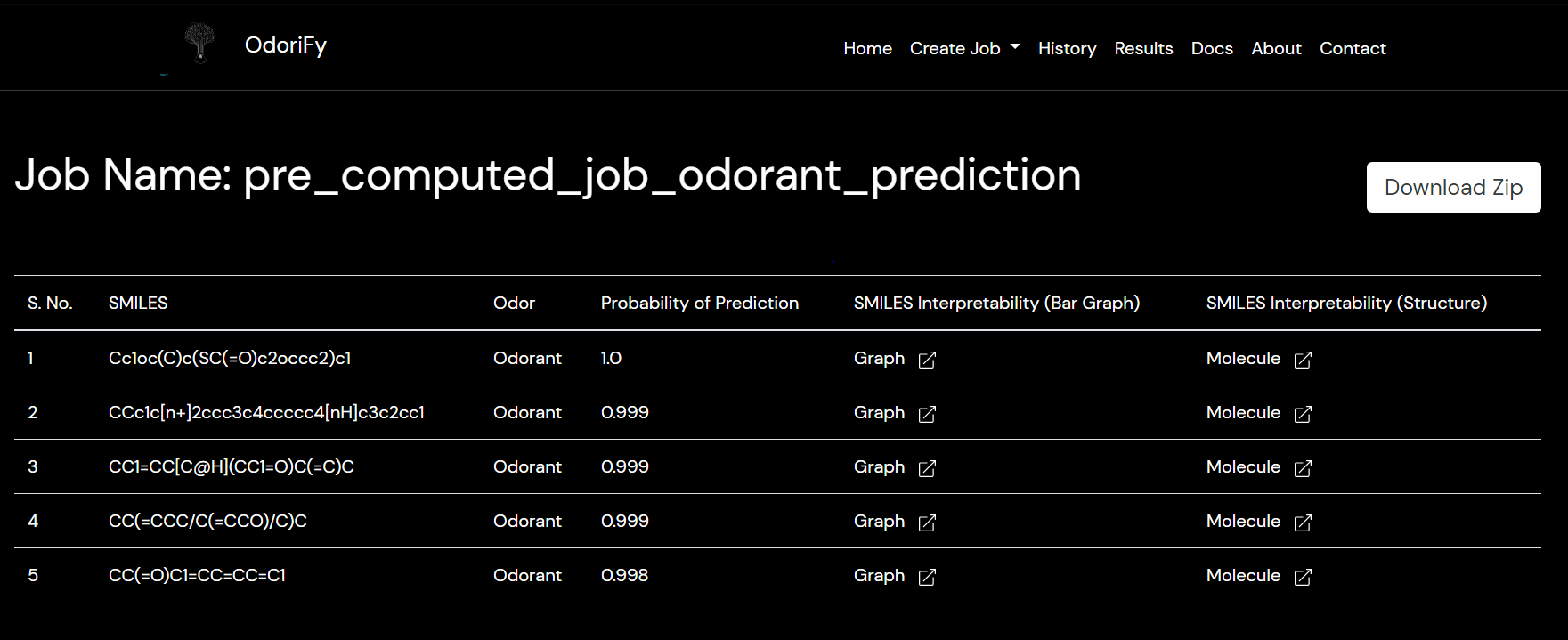
| Prediction Probability | This is the confidence of the prediction being true |
| Receptor Sequence Interpretability | A bar graph representation of relevant amino acids in receptor sequence contributing towards the prediction |
| SMILES Interpretability (Bar Graph) | A bar graph representation of relevant atoms in ligands contributing towards the prediction |
| SMILES Interpretability (Structure) | Substructure Analysis of the ligand (SMILES) highlighting relevant atoms contributing towards the prediction |
Troubleshooting
Troubleshooting
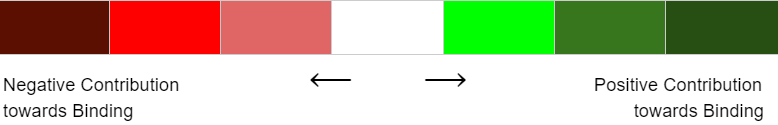
What do the colors in graphs represent?
Q. If I log out of my browser, would my history remain saved?
A. Yes, your history will remain saved up to 7 days, till you choose to clear your cookies in the browser cache.
Q. Can I run 2 prediction models from different tabs of the same browser?
A. No.
Q. Can I navigate away from the loading screen?
A. We understand that it can be a little time consuming, considering the high computations. Please try to be patient, and do not navigate away from the loading screen to get your results. You could, however, add your email address to receive your results.
Q. What if I add more than 25 entries?
A. We only select the first 25 entries as input.
Q. The result page shows a table of only ‘NA’ entries. What does this mean?
A. Don't worry, NA stands for Not Applicable, which indicates that for the given input, there are no ligands/receptors which can bind to the input receptor/ligand with the given input parameters.
Q. How do I interpret my results?
A. The results can be interpreted in three ways: based on Receptor Sequence, based on SMILES & based on Structure Based. The colors in the structure and graphs, green and red represent positive and negative contribution towards the binding, respectively.
Q. I set the counter value of Top-K to be ‘x’, but I receive ‘y’ output records (y<x)?
A. The value of ‘K’ only sets an upper bound of the number of outputs you can get. It is possible to have fewer receptors binding a given input smile than K, or vice versa.
Q. What does the threshold mean?
A. In OR finder, we have used a Tanimoto similarity threshold to find SMILES similar within that threshold. Setting a lower threshold would produce more output records.
Q. How to set a job title? Can I have special characters in my title?
A. Yes, all characters are fit for job titles. We recommend using meaningful job names to keep track of the job. You can see the sample input for more information.
Q. What is the prediction based on?
A. The prediction is based on Deep Learning Models.
Get Started
Get Started
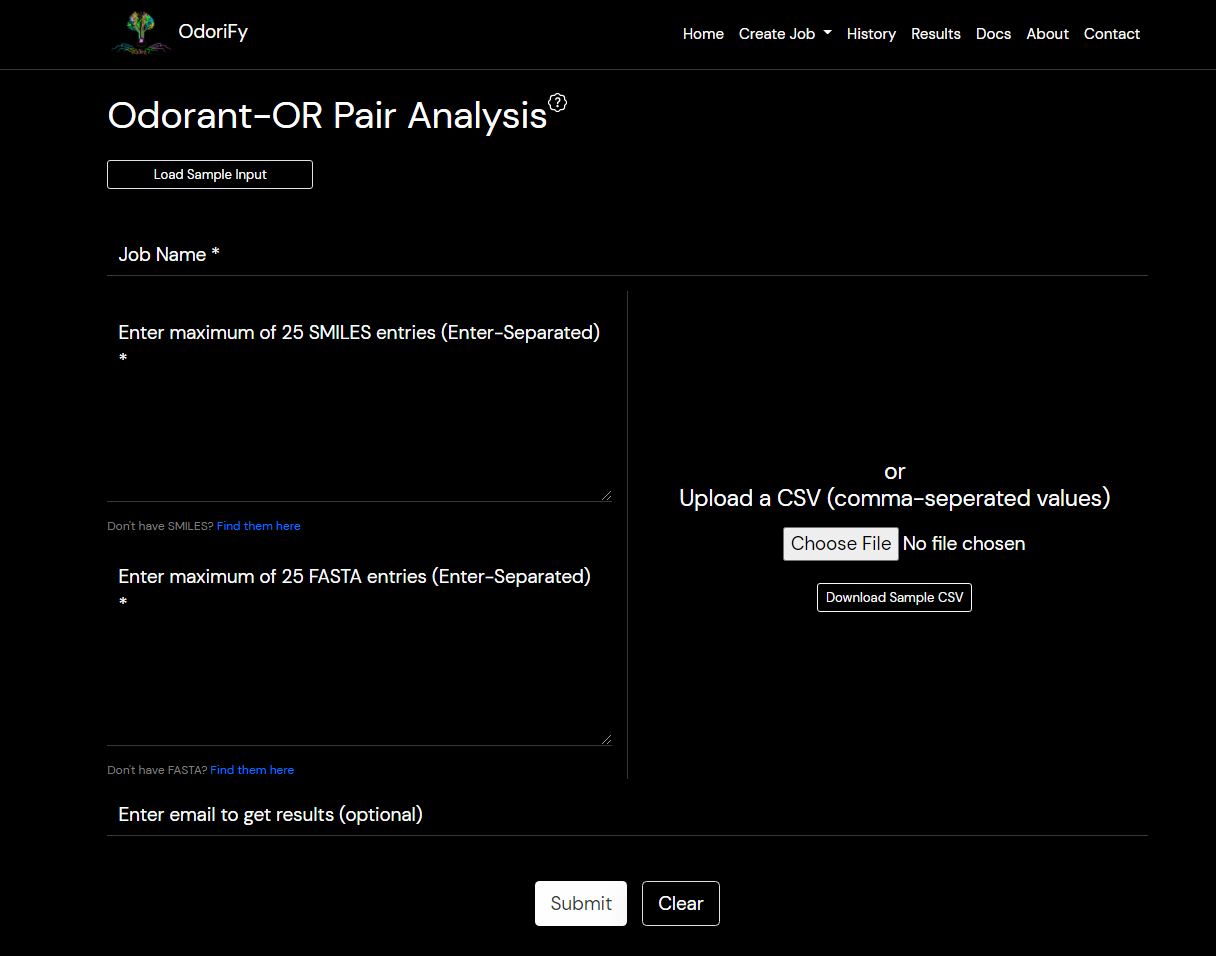
- Go to the Home page. There are 4 prediction engines. Each has a different application and use. They are:-
- Odorant predictor
- OR Finder
- Odor Finder
- Odor-OR Analysis
- Select which prediction engine you need to use according to your requirements. You can choose them on the Prediction Engines page or you can select the engine from the Create Job tab in the menu bar.
- You are required to submit your input files according to the format given below. You can also take a look at the sample input files by downloading them. The sample files are also available on the prediction engines page.
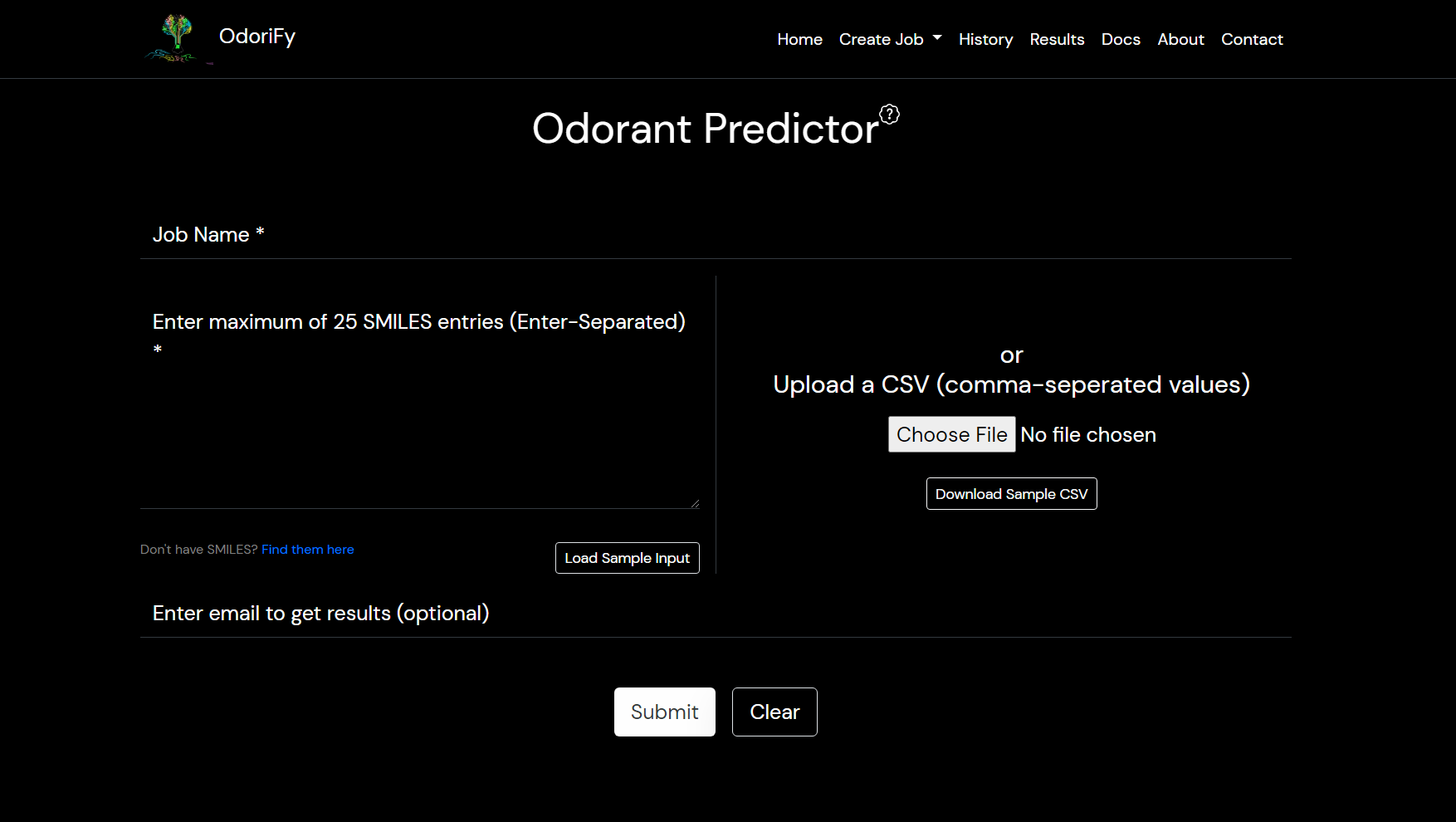
- Odorant predictor: Input is required in the form of SMILES.
There are 2 ways of giving input:- Text box: You have to put ENTER separated SMILES in the box. (You can copy-paste)
- File: A comma-separated values file (.csv file) must be provided containing only a single column of the input smiles.
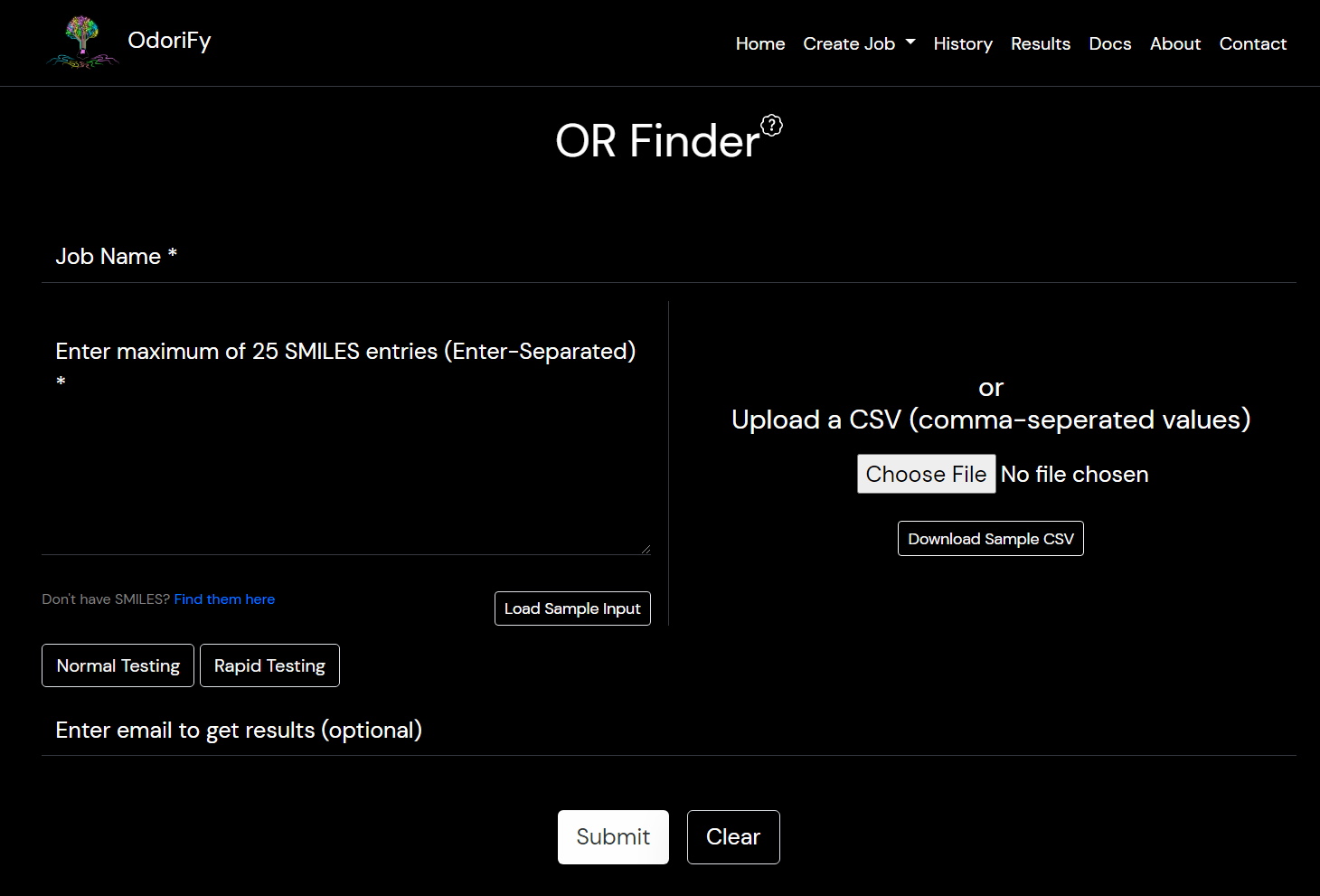
- OR Finder: Input is required in the form of SMILES.
There are 2 ways of giving input:- Text box: You have to put ENTER separated smiles in the box. (You can copy-paste)
- File: A comma-separated values file (.csv file) must be provided containing only a single column of the input smiles.
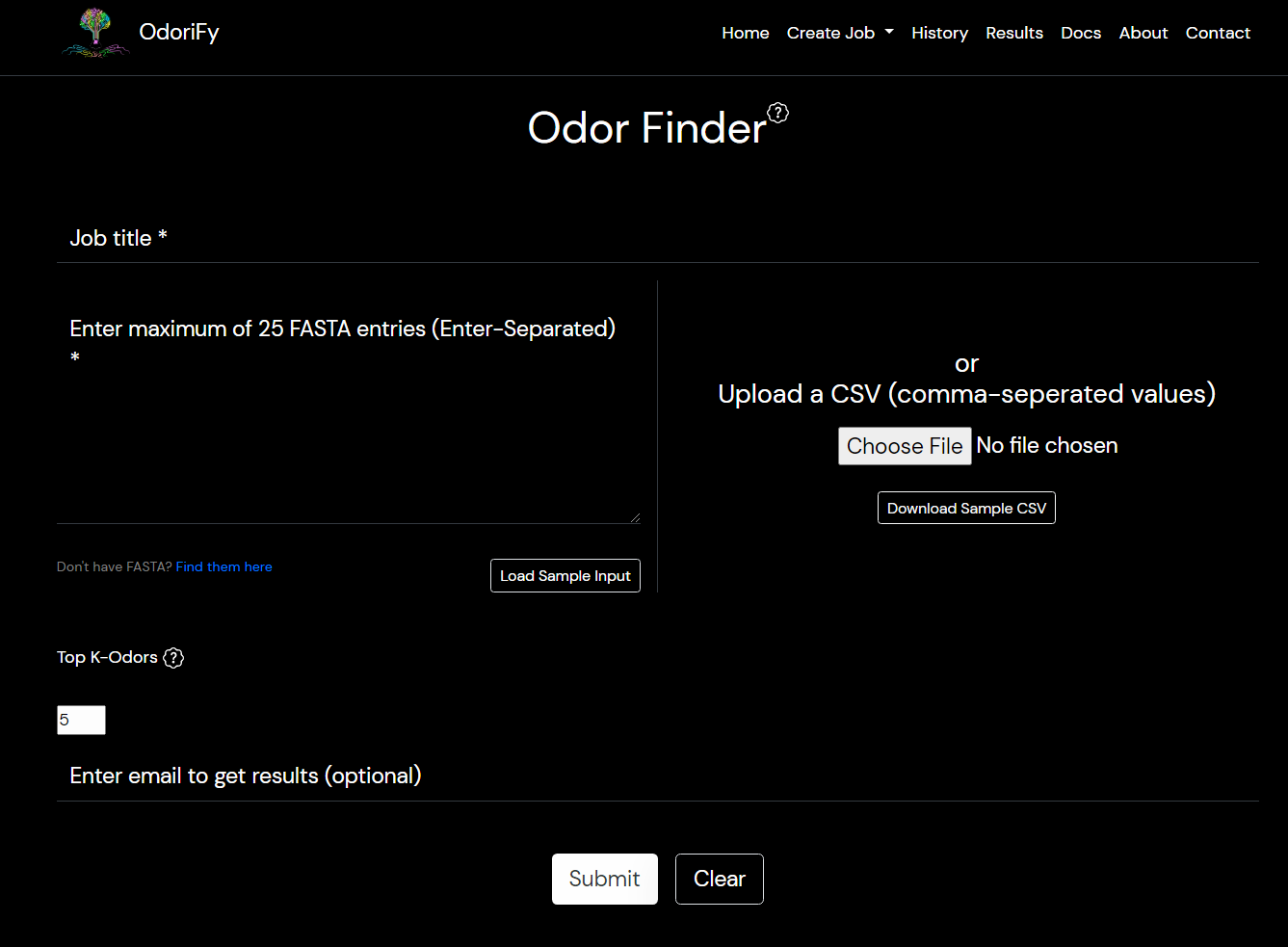
- Odor Finder: Input is required in the form of olfactory receptors FASTA sequence.
There are 2 ways of giving input:- Text box: The format should be the header of FASTA and in the next line the whole receptor sequence. For multiple input sequences, the format should be a FASTA header then next line sequence, then in next line the 2nd input’s header and in next line 2nd input’s sequence and so on.
- File: A comma-separated values file(.csv file) must be provided. 2 columns should be there. 1st column contains the header of the FASTA sequence starting with > and the 2nd column contains the sequence itself.
- Odor-OR Analysis: Input is required in the form of SMILES and protein sequences.
There are 2 ways of giving input:- Text box: You have to put ENTER separated SMILES in box 1. (You can copy-paste), and ENTER separated FASTA Format sequences similar to Odor-Finder in box 2.
- File: A comma-separated values file (.csv file) must be provided with 3 columns. The 1st column should contain the input SMILES.
The 2nd column should contain the corresponding FASTA protein sequence’s header starting with > and the 3rd column should contain the amino acid sequence of the protein.
- Odorant predictor: Input is required in the form of SMILES.
- While submitting the job you will have an option to choose to provide your email, in which case we will send you the results directly on your email and you don’t have to wait after submitting the job.
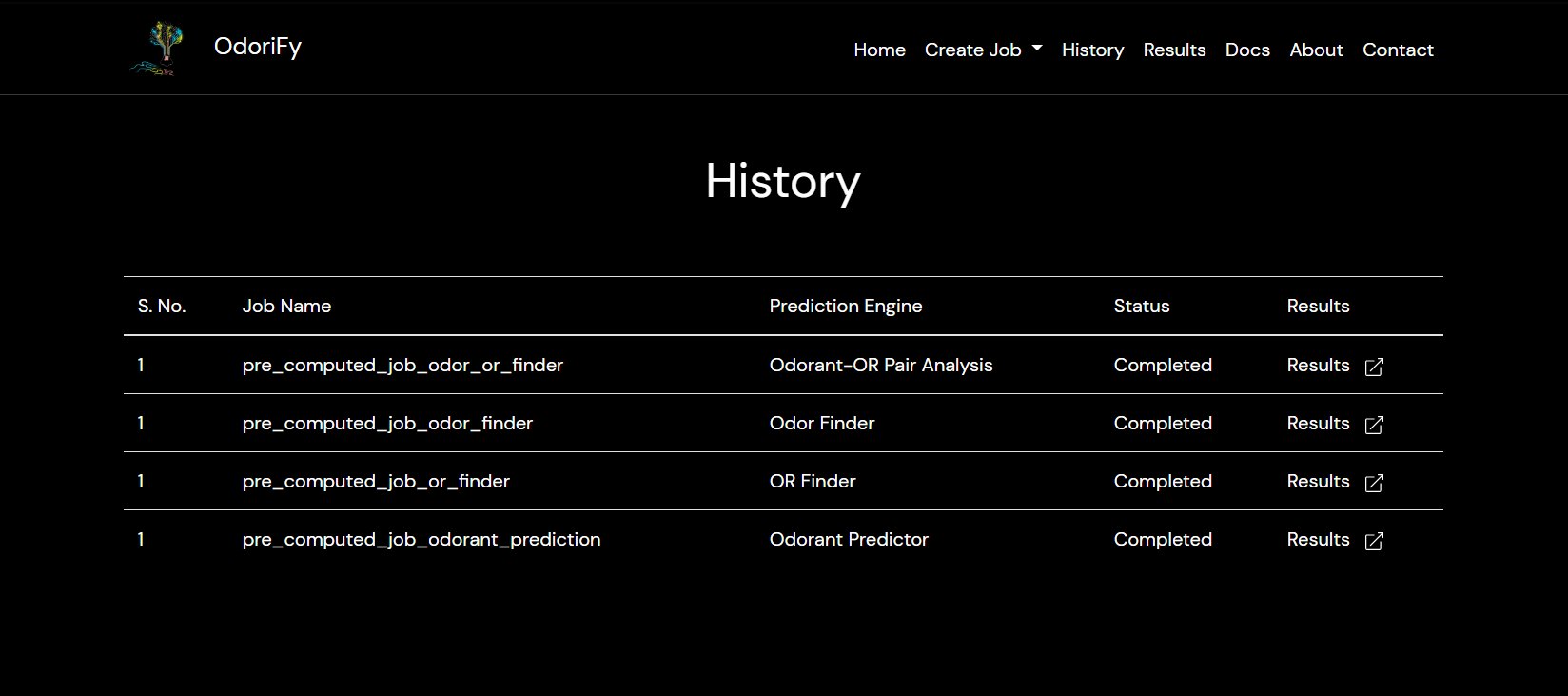
- After the job is completed you will be redirected to the results page where you can see the results of your current job.
- All your jobs will be saved for a week, the results of which can be accessed via the History page.
- In the results page, you also have the option to download the zip file containing all the results for that particular job.